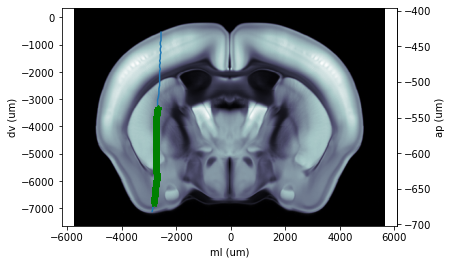
Coronal Plot
Plot a coronal slice (best fit) that contains a given probe track. As input, use an eID and probe label. environment installation guide https://github.com/int-brain-lab/iblenv
[1]:
# Author: Olivier Winter
import numpy as np
from one.api import ONE
import ibllib.atlas as atlas
import brainbox.io.one as bbone
# === Parameters section (edit) ===
eid = 'c7bd79c9-c47e-4ea5-aea3-74dda991b48e'
probe_label = 'probe01'
# === Code (do not edit) ===
ba = atlas.AllenAtlas(25)
one = ONE(base_url='https://openalyx.internationalbrainlab.org')
traj = one.alyx.rest('trajectories', 'list', session=eid,
provenance='Ephys aligned histology track', probe=probe_label)[0]
channels = bbone.load_channel_locations(eid=eid, one=one, probe=probe_label)
picks = one.alyx.rest('insertions', 'read', id=traj['probe_insertion'])['json']
picks = np.array(picks['xyz_picks']) / 1e6
ins = atlas.Insertion.from_dict(traj)
cax = ba.plot_tilted_slice(xyz=picks, axis=1, volume='image')
cax.plot(picks[:, 0] * 1e6, picks[:, 2] * 1e6)
cax.plot(channels[probe_label]['x'] * 1e6, channels[probe_label]['z'] * 1e6, 'g*')
Downloading: C:\Users\Mayo\Downloads\ONE\openalyx.internationalbrainlab.org\zadorlab\Subjects\CSH_ZAD_029\2020-09-19\001\alf\probe01\channels.brainLocationIds_ccf_2017.430ca150-6d52-449d-830e-5339acba3cfe.npy Bytes: 3120
Downloading: C:\Users\Mayo\Downloads\ONE\openalyx.internationalbrainlab.org\zadorlab\Subjects\CSH_ZAD_029\2020-09-19\001\alf\probe01\channels.localCoordinates.2d5dc314-38fd-41d4-b6bb-ba6d232aba9a.npy Bytes: 6064
0%| | 0/3120 [00:00<?, ?it/s]
100%|███████████████████████████████████████████████████████████████████████████| 3120/3120 [00:00<00:00, 69542.49it/s]
100%|██████████████████████████████████████████████████████████████████████████| 6064/6064 [00:00<00:00, 251639.98it/s]
Downloading: C:\Users\Mayo\Downloads\ONE\openalyx.internationalbrainlab.org\zadorlab\Subjects\CSH_ZAD_029\2020-09-19\001\alf\probe01\channels.mlapdv.1bd1b382-1096-4e71-aa35-8459cb4ffacc.npy Bytes: 4616
Downloading: C:\Users\Mayo\Downloads\ONE\openalyx.internationalbrainlab.org\zadorlab\Subjects\CSH_ZAD_029\2020-09-19\001\alf\probe01\channels.rawInd.c2036b4e-4148-46ee-87fa-7abff36a269f.npy Bytes: 3120
0%| | 0/4616 [00:00<?, ?it/s]
100%|██████████████████████████████████████████████████████████████████████████| 4616/4616 [00:00<00:00, 254183.56it/s]
100%|██████████████████████████████████████████████████████████████████████████| 3120/3120 [00:00<00:00, 314792.25it/s]
Out[1]:
[<matplotlib.lines.Line2D at 0x23022c99ac0>]