Loading SpikeSorting Data
Spikesorted output of electrophysiology data.
Relevant Alf objects
channels
clusters
spikes
waveforms
Loading
[2]:
from one.api import ONE
from brainbox.io.one import SpikeSortingLoader
one = ONE(base_url='https://openalyx.internationalbrainlab.org')
[3]:
pid = 'da8dfec1-d265-44e8-84ce-6ae9c109b8bd'
ssl = SpikeSortingLoader(pid=pid, one=one)
spikes, clusters, channels = ssl.load_spike_sorting()
clusters = ssl.merge_clusters(spikes, clusters, channels)
waveforms = ssl.load_spike_sorting_object('waveforms') # loads in the template waveforms
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/spikes.amps.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/spikes.clusters.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/spikes.depths.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/spikes.times.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/clusters.channels.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/clusters.depths.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/clusters.metrics.pqt; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/clusters.uuids.csv; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/channels.brainLocationIds_ccf_2017.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/channels.localCoordinates.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/channels.mlapdv.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/channels.rawInd.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/channels.brainLocationIds_ccf_2017.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/channels.localCoordinates.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/channels.mlapdv.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/channels.rawInd.npy; using most recent
warnings.warn(
Alternatively, one can instantiate the spike sorting loader using the session unique identifier eid and the probe name pname:
[4]:
eid, pname = one.pid2eid(pid)
sl = SpikeSortingLoader(eid=eid, pname=pname, one=one)
spikes, clusters, channels = sl.load_spike_sorting()
clusters = sl.merge_clusters(spikes, clusters, channels)
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/spikes.amps.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/spikes.clusters.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/spikes.depths.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/spikes.times.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/clusters.channels.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/clusters.depths.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/clusters.metrics.pqt; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/clusters.uuids.csv; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/channels.brainLocationIds_ccf_2017.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/channels.localCoordinates.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/channels.mlapdv.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/channels.rawInd.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/channels.brainLocationIds_ccf_2017.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/channels.localCoordinates.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/channels.mlapdv.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/channels.rawInd.npy; using most recent
warnings.warn(
More details
Available spikesorting algorithms
pykilosort (All data)
Kilosort 2.5 (Most data collected before August 2021)
Important information
Data sorted with pykilosort is loaded by default. If the pykilosort spikesorting data is not available, the matlab kilosort 2.5 version will be loaded. See Example 1 for more information.
The channel locations in the brain can come from several sources. it will load the most advanced version of the histology available, regardless of the spike sorting version loaded. The steps, from most to least advanced, are:
alf: the final version of channel locations, same as resolved with the difference that data has been written out to files
resolved: channel location alignments have been agreed upon
aligned: channel locations have been aligned, but review or other alignments are pending, potentially not accurate
traced: the histology track has been recovered from microscopy, however the depths may not match, inacurate data
The attributes
mlapdv,atlas_idsandacronymsin theclustersandchannelsobjects are only available for probe insertions wheresl.histologyis equal to traced, aligned, resolved or alf.The cluster and channel locations in the brain are only considered final for probe insertions with
sl.histology='resolved'orsl.histology='alf'.
Useful modules
Exploring spikesorting data
Example 1: Loading different spikesorting versions
[5]:
# By default, if available, the data spikesorted with pykilosort is loaded.
# To find the spikesorting version that is loaded we can use
sl.collection
# To see all available spikesorted data for this probe insertion we can list the collections.
# N.B. ks2.5 matlab spikesorted data is stored in the alf/probe00 folder
sl.collections
# The following can be used to load a specific version of spikesorting
# pykilosort version
spikes, clusters, channels = sl.load_spike_sorting(spike_sorter='pykilosort')
# ks2.5 matlab version
spikes, clusters, channels = sl.load_spike_sorting(spike_sorter='')
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/spikes.amps.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/spikes.clusters.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/spikes.depths.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/spikes.times.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/clusters.channels.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/clusters.depths.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/clusters.metrics.pqt; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/clusters.uuids.csv; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/channels.brainLocationIds_ccf_2017.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/channels.localCoordinates.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/channels.mlapdv.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/channels.rawInd.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/channels.brainLocationIds_ccf_2017.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/channels.localCoordinates.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/channels.mlapdv.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/channels.rawInd.npy; using most recent
warnings.warn(
Example 2: Loading additional data
[6]:
# The default spikes and cluster attributes loaded are:
# spikes - amps, clusters, depths, times
# cluster - channels, depths, metrics
#Other attributes can additionally be loaded in the following way
spikes, clusters, channels = sl.load_spike_sorting(dataset_types=['clusters.amps', 'spikes.samples'])
clusters = sl.merge_clusters(spikes, clusters, channels)
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/spikes.amps.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/spikes.clusters.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/spikes.depths.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/spikes.samples.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/spikes.times.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2022-10-31#/clusters.amps.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/clusters.channels.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/clusters.depths.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/clusters.metrics.pqt; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/clusters.uuids.csv; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/channels.brainLocationIds_ccf_2017.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/channels.localCoordinates.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/channels.mlapdv.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/channels.rawInd.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/channels.brainLocationIds_ccf_2017.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/channels.localCoordinates.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/channels.mlapdv.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/channels.rawInd.npy; using most recent
warnings.warn(
Example 3: Compute firing rate across session
[7]:
from brainbox.ephys_plots import image_fr_plot
from iblutil.numerical import bincount2D
import numpy as np
time_bin = 0.05 # time bin in seconds
depth_bin = 10 # depth bin in um
# Remove any nan values
kp_idx = np.bitwise_and(~np.isnan(spikes['times']), ~np.isnan(spikes['depths']))
fr, time, depth = bincount2D(spikes['times'][kp_idx], spikes['depths'][kp_idx], time_bin, depth_bin)
Example 4: Find clusters labelled as good
[8]:
good_clusterIDs = clusters['cluster_id'][clusters['label'] == 1]
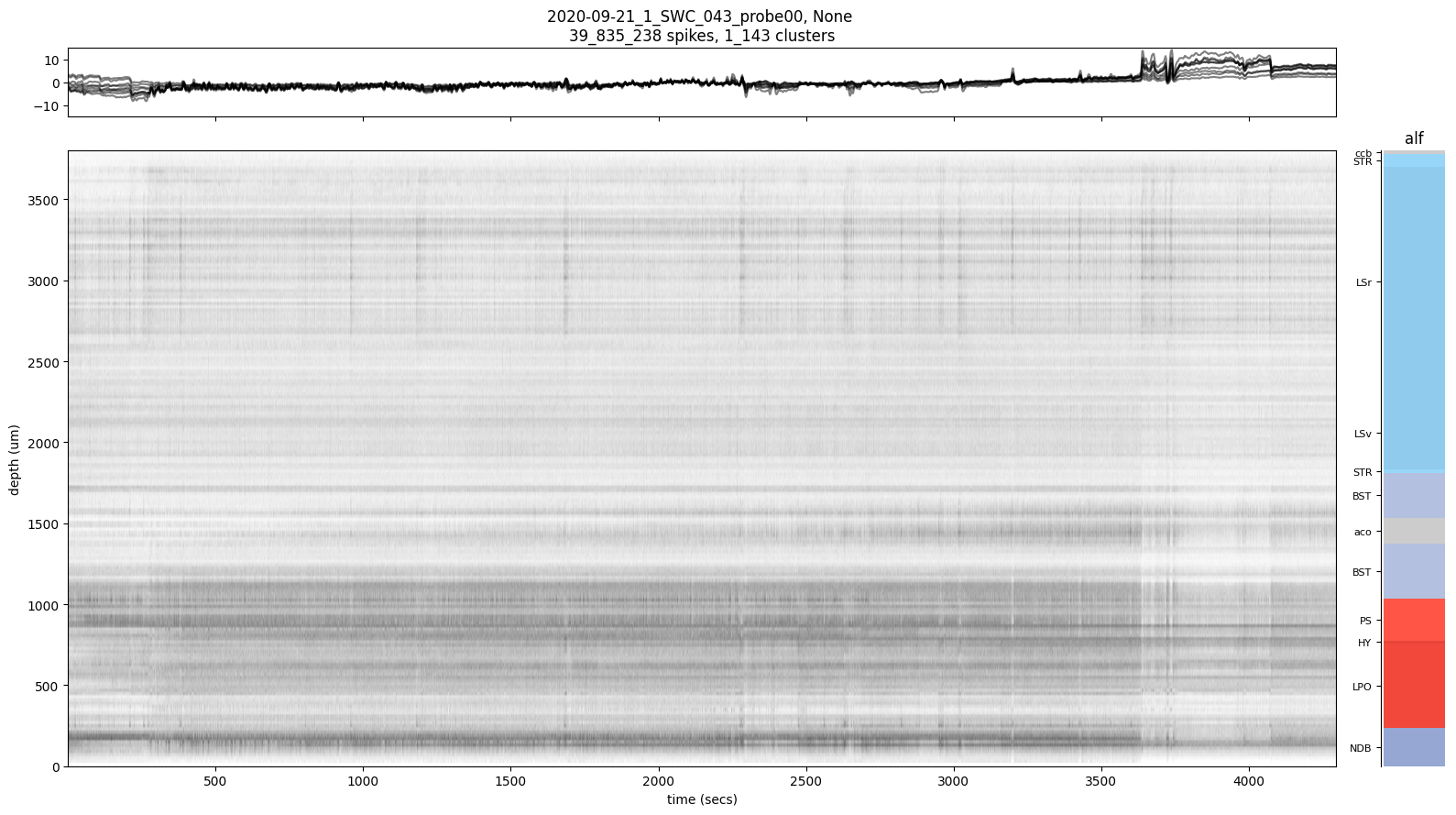
Example 5: Plot a raster for all units
[9]:
sl.raster(spikes, channels)
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/drift.times.npy; using most recent
warnings.warn(
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/one/util.py:406: ALFWarning: No default revision for dataset alf/probe00/pykilosort/#2024-05-06#/drift.um.npy; using most recent
warnings.warn(
Out[9]:
(<Figure size 1600x900 with 4 Axes>,
array([[<Axes: title={'center': '2020-09-21_1_SWC_043_probe00, None \n39_835_238 spikes, 1_143 clusters'}>,
<Axes: >],
[<Axes: xlabel='time (secs)', ylabel='depth (um)'>,
<Axes: title={'center': 'alf'}>]], dtype=object))
Other relevant examples
COMING SOON