Welcome to IBL code library documentation!
IBL data structure
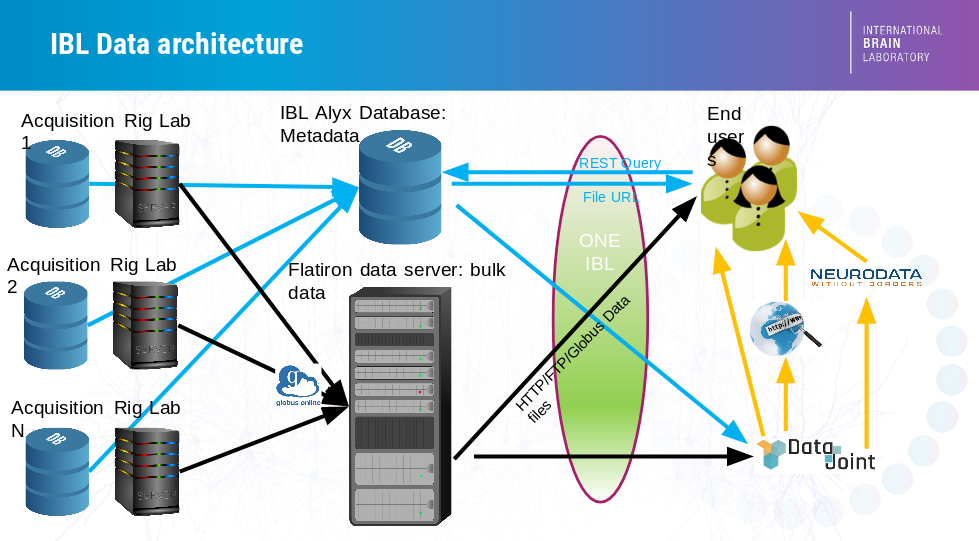
In the IBL, data acquired in laboratories spread across countries needs to be centralized into a common store, accessible from anywhere in the world, at all times. This challenge is met by the IBL data architecture, documented briefly below; a thorough description can be found in our preprint.
The central store has two components:
A Bulk Data Store that stores large raw data files (e.g. raw electrophysiology and video data) as well as pre-processed data (e.g. results of spike sorting or video segmentation). This database is accessible through HTTP, FTP and Globus. This is known informally as the “Flatiron server” as our original data server was generously hosted by the Flatiron Institute.
A Relational Database that stores metadata (e.g. information on each experiment and experimental subject) in a structured manner, together with links to the bulk data files. This database is known as Alyx, for reasons no-one can remember. Alyx contains a web-based front-end to allow users to perform colony management and enter metadata during experiments; documentation on this front end is here. Information on how to connect to Alyx programmatically is here.
Tools to access the data
There are two main ways to access the data:
ONE: an API that connects to the central store, allowing users to search and load data of interest from specific experiments.
Datajoint: a framework to perform automated pipelined analyses on a subset of lightweight data such as behavioral choices and spike times, that allows rapid integration of data from multiple experiments and users.
The full IBL data will be publically released when we have completed collection, preprocessing, curation, and quality control. In the meantime, a subset of curated data are publically available.
Software to analyze IBL data
IBL has released a suite of tools to process and visualize our data.
Brainbox: A library of analysis functions that can be used on IBL data or other neurophysiology recordings.
IBL Viewer: A simple and fast interactive visualization tool based on VTK that uses GPU accelerated volume and surface rendering. From electrophysiological data to neuronal connectivity, this tool allows simple and effective 3D visualization for many use-cases like multi-slicing and time series (even on volumes), and can be embedded within Jupyter Lab/Notebook and Qt user interfaces.
Attention
To get all the software, including ONE, brainbox and visualization tools, install the Unified Environment. This is recommended for IBL members.