ONE Tutorial Matlab¶
Before you begin, make sure you have installed ibllib properly on your system as per the previous instructions.
Create ONE object¶
Once the One class is instantiated and setup, we can create an one object (here labelled one).
With default connection settings¶
Type in Matlab prompt:
one = One(); % this line of code will be the first line to write everytime you re-open Matlab
Reminder: connection parameters inserted via one.setup will modify the JSON .one_params file see installation notes here
With different connection settings for single time use¶
For this tutorial we will be connecting to a test database with a test user. As these credentials will be used for this tutorial only, we do not want to change the base parameters of the JSON .one_params file.
To change the credentials without changing the JSON .one_params file, type:
one = One( 'alyx_login', 'test_user', ...
'alyx_pwd', 'TapetesBloc18', ...
'alyx_url', 'https://test.alyx.internationalbrainlab.org');
You now have created an one object. This object has several fields, the following are of interest for analysis. Type help to retrieve the documentation on each field:
help one.list
help one.load
help one.search
Find an experiment¶
Each experiment is identified by a unique string known as the “experiment ID” (EID). To find an EID, use the one.search command.
The following example shows how to find the experiment(s) performed by the user olivier, on the 24 Aug 2018:
[eid, ses] = one.search('users', {'olivier'}, 'date_range', datenum([2018 8 24 ; 2018 8 24])) ;
returns
eid =
'cf264653-2deb-44cb-aa84-89b82507028a'
The searchable fields are listed by calling the search method with no parameters:
one.search
% Example search keywords:
'dataset_types'
'date_range'
'labs'
'subjects'
'users'
List method¶
Once you know the EID, you can list all the datasets for the experiment using the list command:
one.list(eid)
returns
ans =
29×1 cell array
{'_ibl_lickPiezo.raw' }
{'_ibl_lickPiezo.timestamps' }
{'_ibl_wheel.position' }
{'_ibl_wheel.timestamps' }
{'channels.brainLocation' }
{'channels.probe' }
{'channels.rawRow' }
{'channels.site' }
{'channels.sitePositions' }
{'clusters._phy_annotation' }
{'clusters.depths' }
{'clusters.peakChannel' }
{'clusters.probes' }
{'clusters.templateWaveforms'}
{'clusters.waveformDuration' }
{'eye.area' }
{'eye.blink' }
{'eye.timestamps' }
{'eye.xyPos' }
{'licks.times' }
{'probes.description' }
{'probes.insertion' }
{'probes.rawFilename' }
{'probes.sitePositions' }
{'spikes.amps' }
{'spikes.clusters' }
{'spikes.depths' }
{'spikes.times' }
{'spontaneous.intervals' }
For more detailed datasets info, this command will return a dataclass with dataset_type, data_url and dataset_id fields amongst others:
[dtypes details] = one.list(eid);
details =
struct with fields:
id: {29×1 cell}
name: {29×1 cell}
dataset_type: {29×1 cell}
data_url: {29×1 cell}
data_format: {29×1 cell}
url: {29×1 cell}
eid: {29×1 cell}
To get the full contextual information about the session, get all fields:
ses_info = one.list(eid, 'keyword', 'all')
To navigate further the database, it may be useful to get the range of possible keywords values to search for sessions. For example to print a list of the dataset-types, users and subjects in the command window:
one.list([],'keyword', 'labs')
one.list([],'keyword', 'datasets')
one.list([],'keyword', 'users')
one.list([],'keyword', 'subjects')
Load method¶
General Use¶
To load data for a given EID, use the One.load command:
dataset_types = {'clusters.templateWaveforms', 'clusters.probes', 'clusters.depths'};
eid = 'cf264653-2deb-44cb-aa84-89b82507028a';
[wf, pr, d ]= one.load(eid, 'data' ,dataset_types);
figure,
imagesc(squeeze(wf(1,:,:)), 'Parent', subplot(2,1,1)), colormap('bone')
plot(subplot(2,1,2), squeeze(wf(1,:,:)))
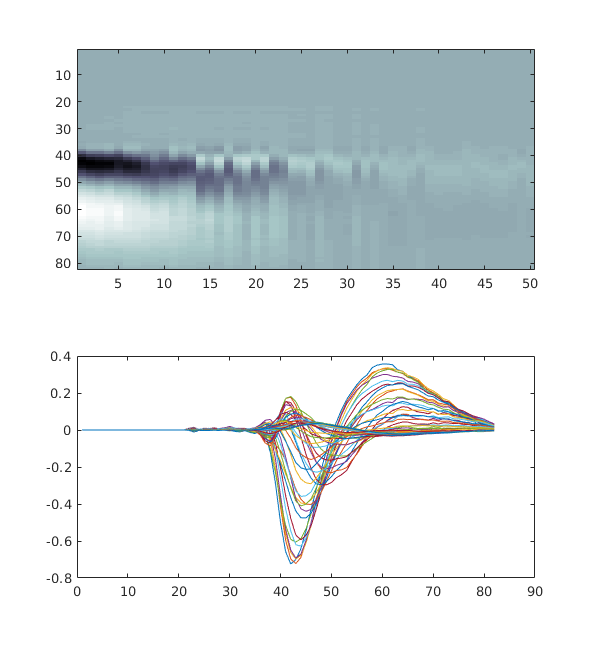
Depending on the use case, it may be handier to wrap the arrays in a dataclass (a structure for Matlab users) so that a bit of context is included with the array. This would be useful when concatenated information for datasets belonging to several sessions.
D = one.load(eid, 'data' ,dataset_types, 'dclass_output', true);
disp(D.local_path)
disp(D.dataset_type)
disp('dimensions: ')
disp(cellfun(@(x) length(x), D.data))
will print in the command window the following:
'/home/owinter/Downloads/FlatIron/mainenlab/Subjects/clns0730/2018-08-24/1/clusters.templateWaveforms.5e7f6ede-2618-41d2-95ee-ce144ea12851.npy'
'/home/owinter/Downloads/FlatIron/mainenlab/Subjects/clns0730/2018-08-24/1/clusters.probes.c41dd877-d511-42cb-90a3-01bb19297117.npy'
'/home/owinter/Downloads/FlatIron/mainenlab/Subjects/clns0730/2018-08-24/1/clusters.depths.42507ace-1ced-4358-a5ef-c4ddd8b7071c.npy'
'clusters.templateWaveforms'
'clusters.probes'
'clusters.depths'
dimensions:
1109
1109
1109
The dataclass contains the following keys, each of which contains a list of 3 items corresponding the the 3 queried datasets
data
cell(*array*): the arraydataset_id
cell(*str*): the UUID of the dataset in Alyxlocal_path
cell(*str*): the local full path of the filedataset_type
cell(*str*): as per Alyx tableurl
cell(*str*): the link on the FlatIron servereid
cell(*str*): the session UUID in Alyx
It is also possible to query all datasets attached to a given session, in which case the output has to be a table/structure as seen above:
[eid, ses_info ]= one.search('subjects', 'flowers');
D = one.load(eid);
In this case it will return a table with 5 datasets.
Specific cases¶
If a dataset type queried doesn’t exist or is not on the FlatIron server, an empty list is returned. This allows to keep the proper order of output arguments
eid = 'cf264653-2deb-44cb-aa84-89b82507028a';
dataset_types = {'clusters.probes', 'thisDataset.IveJustMadeUp', 'clusters.depths'};
[t, empty, cl ] = one.load(eid, 'data', dataset_types)
isempty(empty) % true !
Returns an empty array for empty so that t and cl still get assigned the corresponding datasets values.
Search method¶
The search methods allows to query the database to filter the list of UUIDs according to the following fields:
dataset_typesuserssubjectdate_range
One-to-one matches: subjects¶
This is the simplest case that queries EEIDs (sessions) associated with a subject. There can only be one subject per session.
eid = one.search('subject', 'flowers');
Another use case is to query EEIDs associated with a dataset type.
[eids,ses] = one.search('data', 'channels.brainLocation')
Here is the simple implementation of the filter, where we query for the EEIDs (sessions) co-owned by all of the following users: olivier and niccolo (case-sensitive).
eid = one.search('users',{'test_user', 'olivier'});
The following would get all of the dataset for which olivier is an owner or a co-owner:
[eid, ses_info] = one.search('users', 'olivier');
Note that unlike the first example, here we used an optional second output argument , to get all context information about the returned sessions.
It is also possible to filter sessions using a date-range:
eid = one.search('users','olivier', 'date_range', ['2018-08-24'; '2018-08-24'])
In the example above we used strings to specify the date however it is a safer practice to cast the range as a 2 elements datenum vector.
drange = datenum(['2018-08-24'; '2018-08-24'])
eid = one.search('users','olivier', 'date_range', drange)